In December, scientists identified a SARS-CoV-2 variant of concern (501Y.V2 or B.1.351) in Zambia. Now, just ten weeks later, a CDC report says it appears poised to become the country’s dominant strain.
Moving forward, genetic analyses will be critical in mapping the spread of current and future variants of concern, and in implementing an effective vaccination response.
A nimble genomic response
As the first wave of infections hit Zambia in May 2020, PATH’s Daniel Bridges, PhD, and a team of scientists from the University of Zambia's School of Veterinary Medicine (UNZAVET) quickly got to work sequencing SARS-CoV-2 samples.
Since then, with support from Hokkaido University and the University of Oxford, the team has sequenced close to 300 samples and identified multiple mutations the virus is accumulating.
“There are now hundreds of thousands of publicly available genomes for COVID-19,” says Dr. Bridges. “We are trying to use that data to see how our sequences in Zambia are related and, over time, if there is continued importation into Zambia.”

Dr. Daniel Bridges and Mulenga Mwenda-Chimfwembe analyzing data from a sequencing run. Photo: PATH/Mirriam Chimba.
Samples from individuals infected early in the pandemic were quite diverse with little genetic similarity. That heterogeneity combined with travel information meant the strains were likely imported from Europe and Asia.
“By building up a library of sequences,” says Dr. Bridges, “We are trying to understand whether any of those virus lineages were locally transmitted in Zambia, or if the country’s containment measures were effective in stopping internal spread.”
Uncovering the new variant
While global cases climb into the millions, SARS-CoV-2 has been exploring a huge range of different mutations and combinations. Most of these changes have been “silent” and had little or no effect on the virus, but some mutations have changed how the virus behaves.
In late 2020, sequencing teams in the United Kingdom, South Africa, and Brazil noticed that some virus lineages were expanding far quicker than expected and identified key mutations that made the virus more transmissible.
Once these variants of concern had been identified, the PATH-UNZAVET team started looking for them in recent COVID-19 samples. The team worked day and night—and within six days of receiving samples from the Zambia National Public Health Institute (ZNPHI)—completed the genetic analyses.
From these results, the Zambia Ministry of Health publicly confirmed the presence of the new variant B.1.351, originally identified in South Africa.
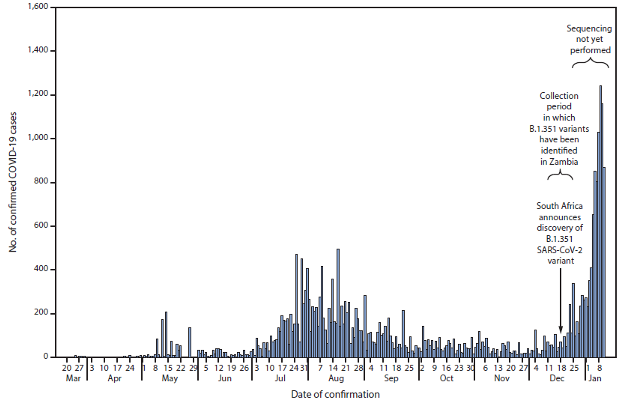
Graph showing the discovery of the B.1.351 variant in Zambia coincides with a sharp increase in COVID-19 cases. Source: Zambia National Public Health Institute.
Seeing the big picture
Now in early 2021, more variants of concern are being identified. These variants may differ from B.1.351—for example, they may be more virulent or have a longer incubation period.
The PATH-UNZAVET team is actively looking for sudden changes in the incidence of a mutation as a clue to what is happening. However, to do that, the team needs to understand how COVID-19 cases are trending.
“We are trying to promote a linkage between the sequencing group and the ZNPHI clinical and surveillance teams so that if they see something different, they get the samples to us to sequence so that we can be more proactive,” says Dr. Bridges.
“We’re looking to better understand how changes at the genetic level correlate to changes in the physical characteristics of the virus. Is it more or less transmissible or virulent? Or able to escape an immune response? That’s the big picture.”
While current evidence suggests the B.1.351 variant is not associated with more severe disease, it is associated with increased transmissibility. Scientists are investigating whether the mutations have any effect on efficacy of therapeutics or vaccines.
Kunda Musonda, MD, PhD, Head of Laboratory Systems and Networks at ZNPHI, emphasizes the connection between this data and national decision-making: “ZNPHI is looking to make the information that is being generated from the research available to policy- and decision-makers.”
The team sees this genomic data as key in getting ahead of the virus and in developing and implementing an effective vaccination response in Zambia and globally.

Dr. Ngonda Saasa, UNZAVET lead researcher, stands next to a PCR machine used to diagnose SARS-CoV-2 infections. Photo: PATH/Mirriam Chimba.
Ngonda Saasa, PhD, a researcher leading UNZAVET activities, notes, “As it is, we are walking behind it and running after it. I wish we could know more about where it comes from, who is more susceptible, how it behaves, and how it changes so that our measures do not become obsolete. The sequencing needs to be done to answer these and many other questions.”
The benefits of this project extend well beyond the Zambia borders. Oxford University’s George Busby, PhD, a collaborator on the project, emphasizes that to maximize the benefits there is a need to ensure data generated in Zambia can be shared with the global effort to sequence SARS-CoV-2.
“There is a lot of unique information that can be collected from genomes,” says Dr. Busby, “and seeing how that data can be worked between different diseases, whether that be work on malaria, TB, COVID-19, and even Ebola or the next virus, is critical. We would like to see this as the start of general traction in the use of this type of data in a whole range of different ways because it does have great potential.”

Joseph Ndebe, UNZAVET laboratory technologist, prepares SARS-CoV-2 samples for sequencing. Photo: PATH/Mirriam Chimba.
Implications beyond the pandemic
COVID-19 has shifted how we prevent, detect, and respond to disease outbreaks. The team is hopeful that these growing genomic capacities could be the start of routine metagenomics for disease surveillance, not just during times of crisis.
Katendi Changula, PhD, a scientist and collaborator at UNZAVET, highlights the value of conducting further research on COVID-19 but warns of not neglecting research on other diseases. “It is still very important to continue other research in zoonosis studies,” says Dr. Changula, “because even if COVID-19 is with us we still have a lot of other diseases that are causing morbidity.”
Building this genomic capacity requires sustained resources. Long-term investment by the Japan International Cooperation Agency and the Japan Agency for Medical Research and Development at UNZAVET will be used to further support the Hokkaido University Research Center for Zoonosis control. By establishing in-country genomic expertise, infrastructure, and responsive platforms, the team hopes to get in front of the current pandemic—and future outbreaks as well.



